A Study of TPL in a Multi-Species Population
The original objective of this project was to investigate a possible connection between Taylor’s Power Law (TPL) and Fisher’s
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from matplotlib.offsetbox import AnchoredText
from scipy import odr
from scipy.interpolate import griddata
from scipy.optimize import newton
from scipy.stats import multivariate_normal
# Plotting config
%matplotlib inline
Domain
Let
Let
If we take
The
Given some
For example, if we are working in a domain with
Because of the combinatorics involved,
def build_phi(dfZ, Q_sup, stat='max', V_scale=10):
assert stat in ('min', 'max', 'mean', 'count')
# Determine ranges and shape of output
N_rng = np.arange(dfZ['N'].min(), dfZ['N'].max() + 1)
assert N_rng[0] > 0
M_rng = N_rng / Q_sup
V_min = dfZ['V'][dfZ['V'] > 0].min() # remove zero variance
V_rng = np.linspace(V_min, dfZ['V'].max(), len(N_rng) * V_scale)
shape = (len(M_rng), len(V_rng))
# This matrix counts number of data point in each 2d bin
bin_counts, _, _ = np.histogram2d(
dfZ['M'], dfZ['V'],
bins=shape,
range=[
[M_rng[0], M_rng[-1]],
[V_rng[0], V_rng[-1]]],
density=False,
)
# This matrix sums up the given statistic for each 2d bin
Phi, _, _ = np.histogram2d(
dfZ['M'], dfZ['V'],
weights=dfZ[stat],
bins=shape,
range=[
[M_rng[0], M_rng[-1]],
[V_rng[0], V_rng[-1]]],
density=False,
)
# Divide the stat sums by their corresponding counts
Phi[bin_counts>0] /= bin_counts[bin_counts>0]
# Round up or down for "max" and "min" stats
if stat == 'max':
Phi = np.ceil(Phi).astype(int)
elif stat == 'min':
Phi = np.floor(Phi).astype(int)
return Phi, N_rng, M_rng, V_rng
# Phi for Q=50 and 1<=N<=100
dfZ_a = pd.read_hdf('zeros_q50_n100.h5', 'df')
Phi_a, N_rng_a, M_rng_a, V_rng_a = build_phi(dfZ_a, 50, 'mean')
# Phi for Q=100 and 1<=N<=100
dfZ_b = pd.read_hdf('zeros_q100_n100.h5', 'df')
Phi_b, N_rngb, M_rng_b, V_rng_b = build_phi(dfZ_b, 100, 'mean')
# Plot
fig, ax = plt.subplots(1, 2, sharey=False)
fig.set_figwidth(12)
ax[0].pcolor(M_rng_a, V_rng_a, Phi_a.T, cmap='magma')
ax[0].set_title('$\Phi$ for $Q=50$')
ax[0].set_xlabel('Mean')
ax[0].set_ylabel('Variance')
ax[1].pcolor(M_rng_b, V_rng_b, Phi_b.T, cmap='magma')
ax[1].set_title('$\Phi$ for $Q=100$')
ax[1].set_xlabel('Mean')
ax[1].set_ylabel('Variance');
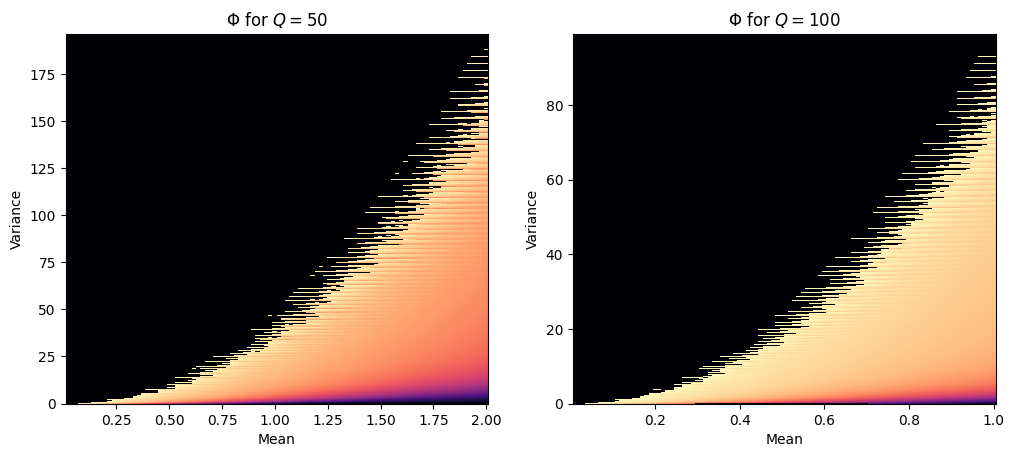
The two heatmaps above show
Random populations
def orth_fit(x, y):
odr_obj = odr.ODR(odr.Data(x, y), odr.unilinear)
output = odr_obj.run()
return output.beta
def calc_rho(pdf, Phi, Q, Q_sup):
pdf = pdf.T.copy()
pdf[Phi<=0] = 0 # get rid of impossible values
pdf /= pdf.sum()
# For each (M,V), if Phi(M,V) zeros are expected (of Q_sup bins),
# then to the probability of getting Q zeros in a row at the beginning
# is this product.
lam = 1
for i in range(Q):
rem = Phi-i
rem[rem<0] = 0
lam *= rem / (Q_sup-i)
rho = (pdf * lam).sum()
return rho
class Population:
def __init__(self, path, Q_sup=None, N_sup=None, S_sup=None, name=None):
self.Q_sup = 50 if Q_sup is None else Q_sup
self.N_sup = 100 if N_sup is None else N_sup
self.S_sup = 200 if S_sup is None else S_sup
self.name = 'Population' if name is None else name
self.Q_cols = np.array([f'Q{i}' for i in range(self.Q_sup)]) # column names
self.df_path = path
self.df = pd.read_hdf(self.df_path, 'df')
self.dfZ_path = f'zeros_q{self.Q_sup}_n{self.N_sup}.h5'
self.dfZ = pd.read_hdf(self.dfZ_path, 'df')
self.Phi, self.N_rng, self.M_rng, self.V_rng = build_phi(self.dfZ, self.Q_sup, 'max')
self.pdf_true = self.get_pdf(self.df['M'].to_numpy(), self.df['V'].to_numpy())
self.dfS = self.simulate()
def get_pdf(self, M, V, allow_singular=False):
# Fit a 2D normal distribution to the data (in log space)
M_ln = np.log(M)
V_ln = np.log(V)
rv = multivariate_normal(
mean=[np.mean(M_ln), np.mean(V_ln)],
cov=np.cov(M_ln, V_ln),
allow_singular=allow_singular,
)
Xm, Ym = np.meshgrid(self.M_rng, self.V_rng)
pos = np.dstack((Xm, Ym))
pdf = rv.pdf(np.log(pos))
pdf /= pdf.sum()
pdf[np.isclose(pdf, 0)] = 0
pdf /= pdf.sum()
return pdf
def simulate(self, shuffle=False):
# Select bin data
Q_cols = self.Q_cols.copy()
if shuffle:
np.random.shuffle(Q_cols)
df = self.df[Q_cols].copy()
# Count unique species across entire domain, starting from left bin
dfS = pd.DataFrame({
'Q': range(1, len(Q_cols)+1),
'S': np.count_nonzero(
np.cumsum(df, axis=1),
axis=0),
})
for i in range(len(Q_cols)):
cols = Q_cols[:i+1]
_df = df[cols].copy()
_N = np.sum(_df, axis=1)
_M = np.mean(_df, axis=1)
_V = np.var(_df, axis=1)
_df['N'] = _N
_df['M'] = _M
_df['V'] = _V
# Discard species with zero mean or variance
_df = _df[(_df['M'] > 0) & (_df['V'] > 0)]
tpl_b, tpl_a, rho = np.nan, np.nan, np.nan
if len(_df.index) > 1:
M_ln = np.log(_df['M'].to_numpy())
V_ln = np.log(_df['V'].to_numpy())
tpl_b, tpl_a_ln = orth_fit(M_ln, V_ln)
tpl_a = np.exp(tpl_a_ln)
# Note: using true PDF
pdf = self.pdf_true
rho = calc_rho(pdf, self.Phi, i+1, self.Q_sup)
dfS.loc[i, ['b', 'a', 'rho']] = tpl_b, tpl_a, rho
return dfS
# Population datasets
populations = [
Population('pop_4_q50_n100_s200.h5', Q_sup=50, name='A1'),
Population('pop_5_q50_n100_s200.h5', Q_sup=50, name='A2'),
Population('pop_6_q50_n100_s200.h5', Q_sup=50, name='A3'),
Population('pop_1_q100_n100_s200.h5', Q_sup=100, name='B1'),
Population('pop_2_q100_n100_s200.h5', Q_sup=100, name='B2'),
Population('pop_3_q100_n100_s200.h5', Q_sup=100, name='B3'),
]
def scatter_plot(pop, ax=None):
if ax is None:
fig, ax = plt.subplots(1, 1)
M = pop.df['M']
V = pop.df['V']
M_ln = np.log(M)
V_ln = np.log(V)
N_rng = pop.N_rng
M_rng = pop.M_rng
ax.scatter(M, V, 1, marker='+')
ax.set_xscale('log')
ax.set_yscale('log')
ax.set_title(f'Population {pop.name}')
# ax.set_xlabel('M')
# ax.set_ylabel('V')
V_bounds = np.zeros((len(M_rng), 2))
for i, n in enumerate(N_rng):
s = np.zeros(pop.Q_sup)
s[0] = n
max_ = s.var()
quo, mod = np.divmod(n, pop.Q_sup)
s[:mod] = quo + 1
s[mod:] = quo
assert s.sum() == n
min_ = s.var()
V_bounds[i] = (min_, max_)
ax.plot(M_rng, V_bounds[:, 0], '--', color='tab:gray')
ax.plot(M_rng, V_bounds[:, 1], '--', color='tab:gray')
est_b, est_a_ln = orth_fit(M_ln, V_ln)
est_a = np.exp(est_a_ln)
V_tpl = np.exp(np.log(est_a)+est_b*np.log(M))
ax.plot(M, V_tpl, 'b-', linewidth=1)
# ax = plt.gca()
tpl_text = '$b = %.2f$ $a = %.2f$' % (est_b, est_a)
at = AnchoredText(
tpl_text, loc='upper left', prop=dict(size=8), frameon=True)
at.patch.set_boxstyle('round, pad=0., rounding_size=0.2')
ax.add_artist(at)
# Plot
fig, ax = plt.subplots(2, 3, sharey=True)
fig.set_figheight(8)
fig.set_figwidth(16)
plt.subplots_adjust(hspace=0.4)
for i, pop in enumerate(populations):
row = int(i/ax.shape[1])
col = i % ax.shape[1]
scatter_plot(pop, ax[row][col])
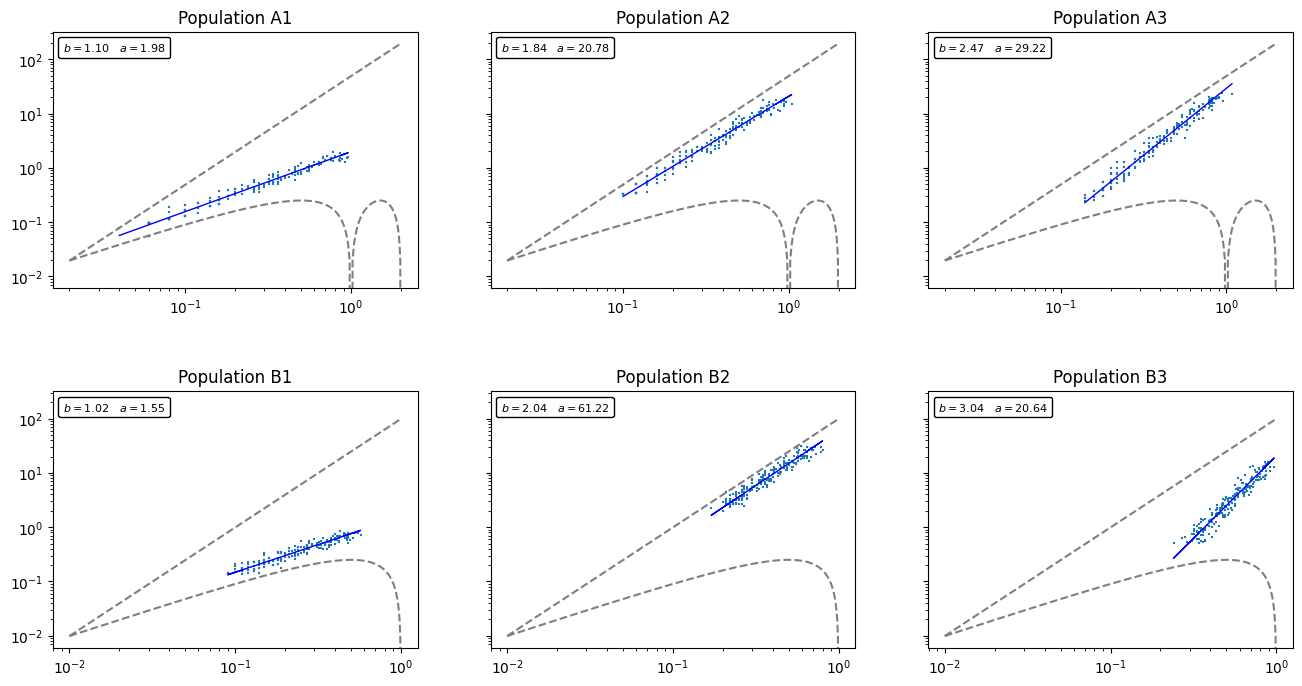
I’ve generated 6 random multi-species populations. The first 3 (labels starting with A) assume B) assume
The question I’m interested in: How do the distributions of these different populations affect the expected species count
It turns out that when a population consists mainly of highly aggregated species, then it will take longer for
As an illustration, let’s plot out
for i in [3, 4]:
pop = populations[i]
plt.plot(pop.dfS['Q'], pop.dfS['S'], label=f'Population {pop.name}')
plt.legend()
plt.xlabel('$q$')
plt.ylabel('$s_q$')
plt.title('Convergence of $s_q \\rightarrow S$ as $q \\rightarrow Q$');
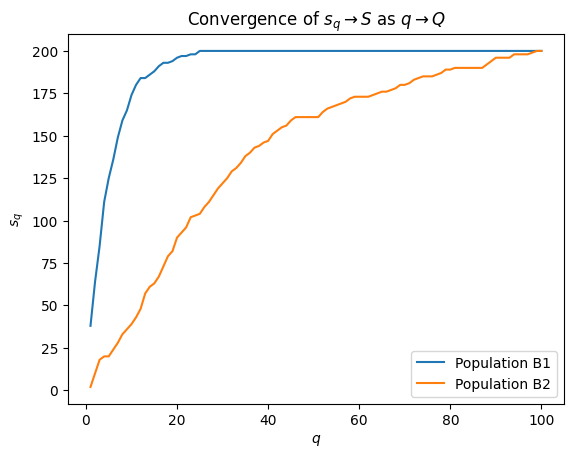
Notice the clear difference in shape between the convergence lines of populations B1 and B2. Population B1 has less variance than B2 (therefore more uniform), so we can expect the various species to be discovered at earlier iterations
So, in theory, if you know how much as been sampled and you have a good idea as to the degree of aggregation in the population, then you should be able to place an estimate on the absolute total number of species.
Estimating
Let
Recall from before that the probability that a species with a given
If
We are essentially piece-wise multipying
Rewritten we have a formula for estimating
The key concept is the estimation of
lam = 1
q = 20
for i in range(q):
rem = Phi_b-i
rem[rem<0] = 0
lam *= rem / (pop.Q_sup-i)
pop = populations[4] # B2
fig, ax = plt.subplots(1, 3, sharey=True)
fig.set_figwidth(16)
ax[0].pcolor(M_rng_b, V_rng_b, pop.pdf_true, cmap='magma')
ax[0].set_title('PDF')
ax[0].set_ylabel('V')
ax[0].set_xlabel('M')
ax[1].pcolor(M_rng_b, V_rng_b, lam.T, cmap='magma')
ax[1].set_title('$\lambda_{%i}$' % q)
ax[1].set_xlabel('M')
ax[2].pcolor(M_rng_b, V_rng_b, pop.pdf_true * lam.T, cmap='magma')
ax[2].set_title('PDF$\cdot\lambda_{%i}$' % q)
ax[2].set_xlabel('M');
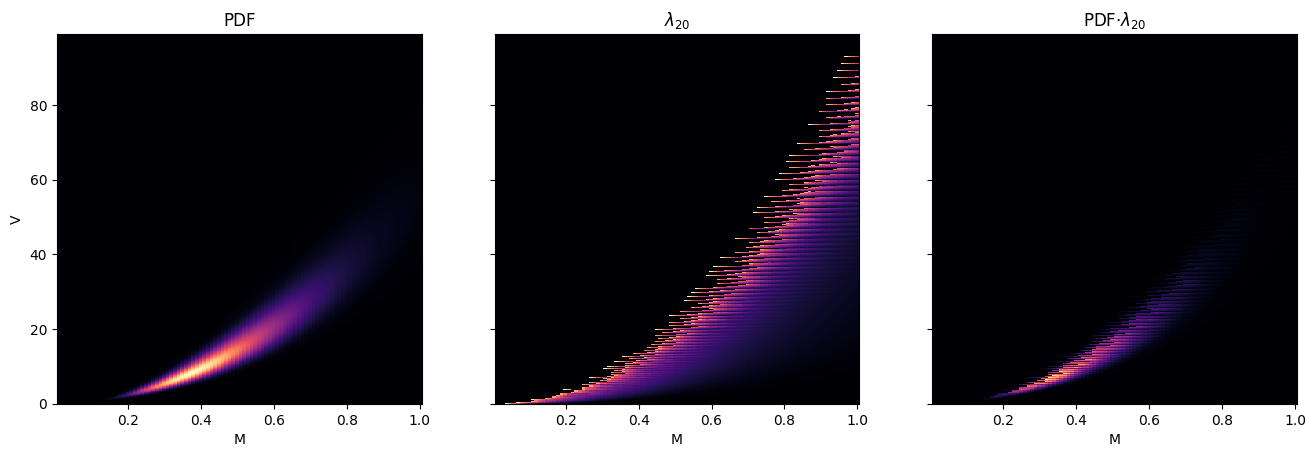
The first plot is the PDF of Population B2 in
Results
Now let’s test the theory for each of the 6 populations.
def sim_plot(pop, ax=None, shuffle=False):
if ax is None:
fig, ax = plt.subplots(1, 1)
dfS = pop.simulate(shuffle=True) if shuffle else pop.dfS
ax.plot(dfS['Q'], dfS['S'])
ax.plot(dfS['Q'], pop.S_sup * (1-dfS['rho']))
ax.plot(dfS['Q'], dfS['S'] / (1-dfS['rho']))
ax.set_title(f'Population {pop.name}')
ax.set_xlabel('q')
# Cumulative species count
fig, ax = plt.subplots(2, 3, sharey=True)
fig.set_figheight(8)
fig.set_figwidth(16)
plt.subplots_adjust(hspace=0.4)
for i, pop in enumerate(populations):
row = int(i/ax.shape[1])
col = i % ax.shape[1]
sim_plot(pop, ax=ax[row][col], shuffle=False)
ax[0][0].set_ylabel('species count')
ax[1][0].set_ylabel('species count');
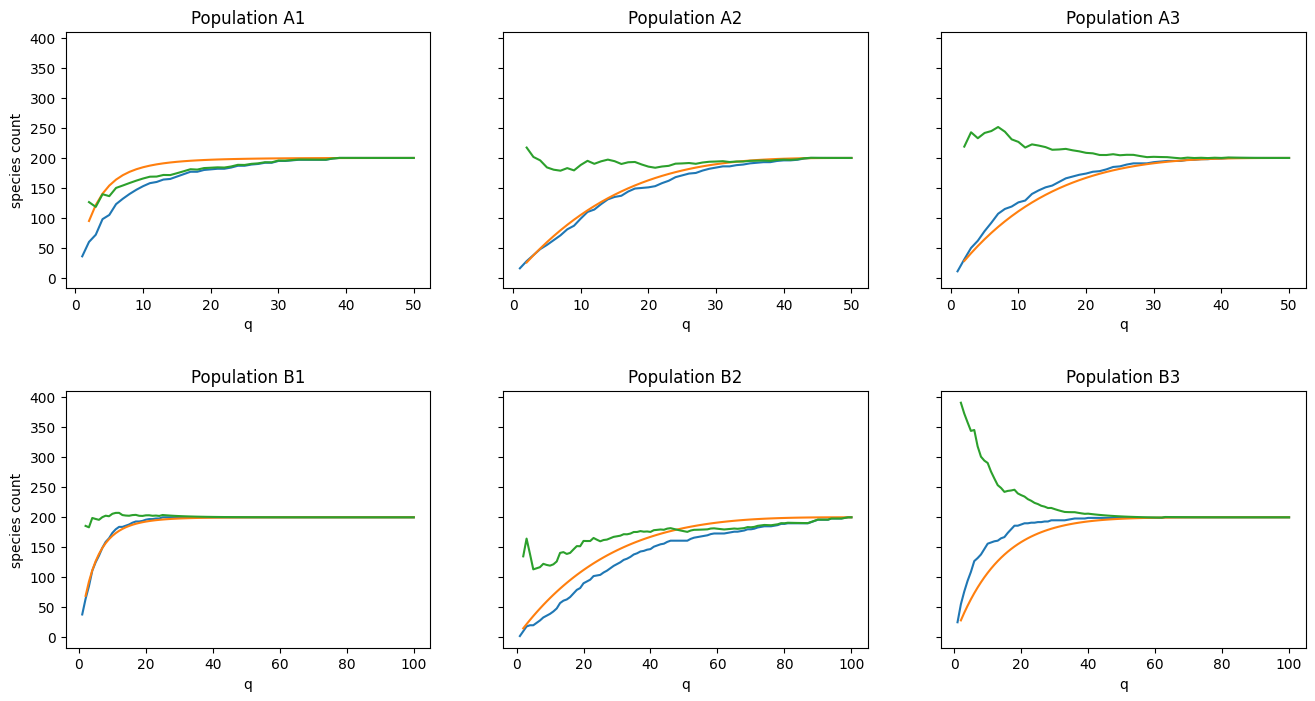
How to interpret these plots:
- The blue line is the observed number of species
as . - The green line is our estimate of
given (we expect this line to approach in all cases). - The orange curve is what we expect our species curve to look like given
and prior knowledge of . The closer the blue and orange lines are together, the better our estimate of will be (green line).
Visual inspection yields mixed results:
- Populations A2, A3, and B1 show excellent alignment
- Populations A1 and B2 show “okay” alignment
- Population B3 shows very poor alignment
Discussion
First of all, the procedure for estimating
Secondly, it can be seen clearly from the results that the procedure is not fool-proof. In particular, predictions for Populations A1 and B2 were farther off than I’d like, while B3 was way off. My hunch is that these outliers are due to a failure to take into account the higher-order moments of
It may be possible to side-step both issues by applying a non-linear machine learning algorithm (e.g. neural network) to the available data. There is clearly a strong connection between the population distribution,
Now let’s consider the
Possible lead?
Is it possible to somehow remove the need for prior knowledge of
Given:
- Let
be the PDF of all species in space after all samples have been taken. - Let
be the PDF of observed species after samples taken. - Let
be the PDF of unobserved species after samples taken.
It follows that
We can write
If
Thus we have a relation between
Here we have a relation between two unknowns. There’s not much more to be done without additional traction. Perhaps this can be formulated as an optimization problem in which we simultaneously solve for
Here’s a thought experiment. Let’s say you’ve taken
So, given our first sample, right off the bat there are certain
References
- Taylor, R. A. (2019). Taylor’s power law: order and pattern in nature. Academic Press.
- Fisher, R. A., Corbet, A. S., & Williams, C. B. (1943). The relation between the number of species and the number of individuals in a random sample of an animal population. The Journal of Animal Ecology, 42-58.
